myCSUSM
myCSUSMFine-tuning convolutional neural networks to classify marine protists
Author 1, Author 2, Author 3, Author 4
Introduction + Goal
- Plankton make up the base of the food web and populations respond rapidly to changes in the environment.
- Lingulodinium polyedrum is a mixotrophic dinoflagellate, forms bioluminescent blooms in southern California.
- Ciliates are heterotrophic protists, they consume and graze on plankton.
- These organisms are hard to study because they are microscopic, and they are abundant making them time consuming to study. They also lack pigment so they cannot be rapidly detected by pigments.
- Teaching a computer to classify images can automate this process and cut down on classification time.
- We want to study these organisms to gain insight on their interaction between one another.
- Goal is to train and use a machine learning system called a convolutional neural network (CNN), to classify images to classify plankton before and during bloom.
Methods
- Collection of Images: A camera is located off of Scripps Memorial Pier in La Jolla,
CA.
- Purpose is to collect continuous images of plankton .
- Organization of Data: Images were obtained from the Scripps pier location and were classified by a human into 4 categories: (Ciliate, L_poly, Questionable, Other.)
- All images were quality controlled by humans. First, images were put into a questionable
folder in order to ensure that all images that were being classified were 100% accurate,
images in the questionable category were analyzed later by Dr. Taniguchi and other
students. We also had at least two people label the same set of images in order to
ensure that the images being classified belong to the correct classification.
- Part 1: We labeled about 258,660 images from every tuesday of every month in 2018
and 2019.
- The purpose of this is to train an existing CNN (Convolutional Neural Network) and fine tuning it to classify plankton.
- Part 2 : The second included the labeling of images from two dates in 2020 (One day
was before the bloom in February and the other date was during the bloom in April.)
- Purpose: We are trying to see how well the classifier does with new novel data.**** With this information we will be able to calculate the error rate.
- Quality controlling the images: images were labeled by at least two people in order
to ensure that the images being classified belong to the correct classification.
- Purpose: The overall goal is to train the existing CNN to get more accurate high quality results.
- Part 1: We labeled about 258,660 images from every tuesday of every month in 2018
and 2019.
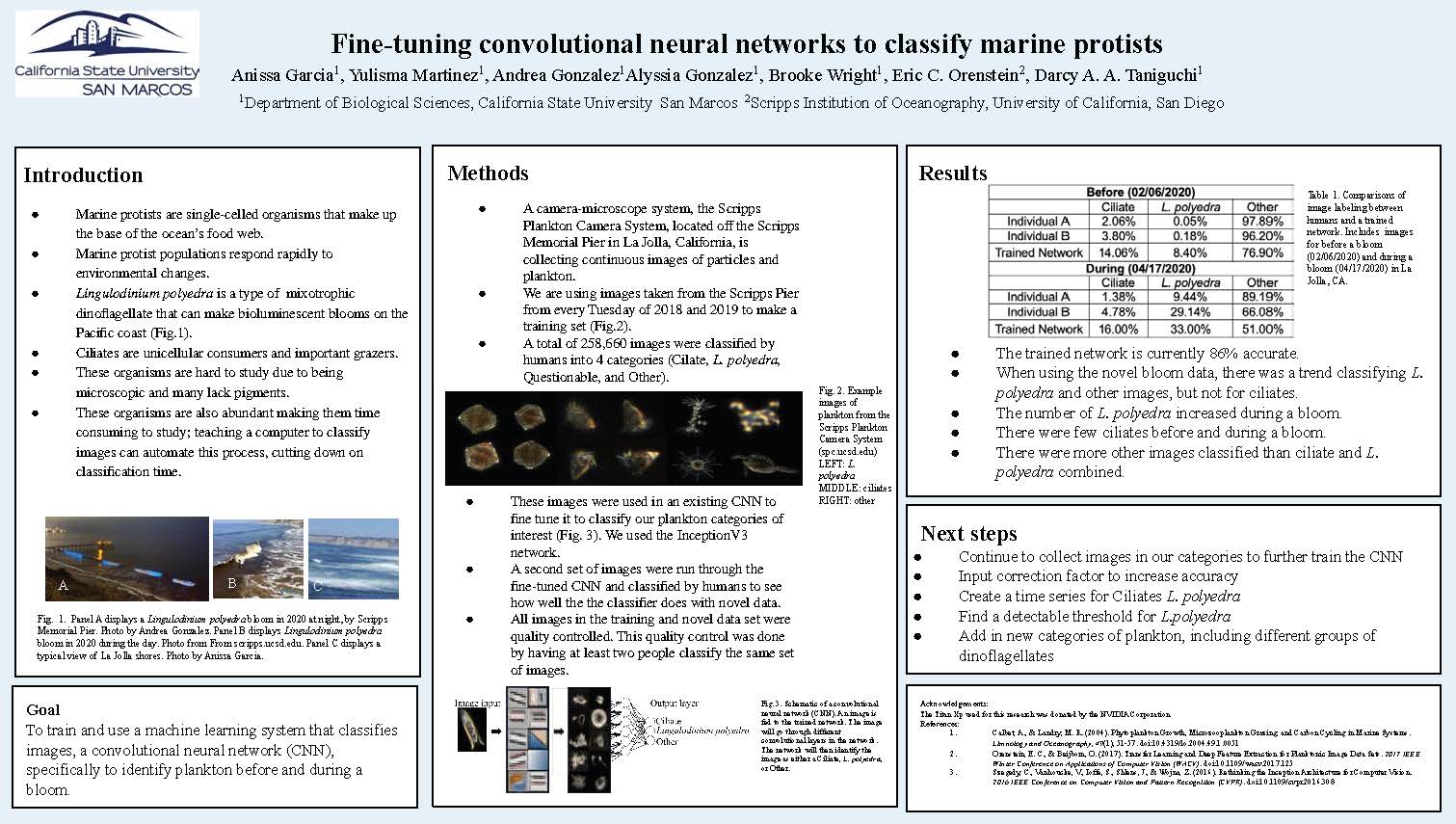
Results
| Ciliate | L. polyedra | Other | |
|---|---|---|---|
| Individual A | 2.06% | 0.05% | 97.89% |
| Individual B | 3.80% | 0.18% | 96.20% |
| Trained Network | 14.06% | 8.40% | 76.90% |
| Ciliate | L. polyedra | Other | |
|---|---|---|---|
| Individual A | 1.38% | 9.44% | 89.19% |
| Individual B | 4.78% | 29.14% | 66.08% |
| Trained Network | 16.00% | 33.00% | 51.00% |
Table 1 & 2 Summary: Comparisons of image labeling between humans and a trained network. Includes images for before a bloom (02/06/2020) and during a bloom (04/17/2020) in La Jolla, CA.
- The trained network is currently 86% accurate.
- When using the novel bloom data, there was a trend classifying L. polyedra and other images, but not for ciliates.
- The number of L. polyedra increased during a bloom.
- There were few ciliates before and during a bloom.
- There were more other images classified than ciliate and L. polyedra combined.
Next steps
- In order to further this study we would like to continue to collect images in our chosen categories to train CNN. We will also quality control these images to ensure that we get the best results possible.
- Input correction factor to increase accuracy. We want the highest accuracy possible.
- Create a time series for these organisms-before, during, after bloom (Ciliates/ polyedra). With this we would be able to observe how these organisms progress through time and may be able to determine if there is a relationship among them.
- Find a detectable threshold for polyedra so we can alert people of when a bloom is beginning.
- Add in new categories of plankton, including different groups of dinoflagellates
Acknowledgements
The Titan Xp used for this research was donated by the NVIDIA Corporation
References
- Calbet, A., & Landry, M. R. (2004). Phytoplankton Growth, Microzooplankton Grazing, and Carbon Cycling in Marine Systems. Limnology and Oceanography, 49(1), 51-57. doi:10.4319/lo.2004.49.1.0051
- Orenstein, E. C., & Beijbom, O. (2017). Transfer Learning and Deep Feature Extraction for Planktonic Image Data Sets. 2017 IEEE Winter Conference on Applications of Computer Vision (WACV). doi:10.1109/wacv.2017.125
- Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., & Wojna, Z. (2016). Rethinking the Inception Architecture for Computer Vision. 2016 n IEEE Conference on Computer Vision and Pattern Recognition (CVPR). doi:10.1109/cvpr.2016.308